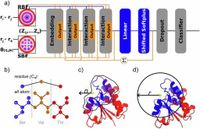
In recent years, the understanding of enzyme function and classification has undergone significant evolution, with computational models leading the charge. The advent of tools that infer enzyme function from sequence and structure information has revolutionized the field; however, challenges persist due to misclassifications that arise when local structural nuances influence enzyme activity. Addressing this crucial issue, researchers have now introduced TopEC, a state-of-the-art 3D graph neural network (GNN) designed specifically to predict enzyme classifications by analyzing enzyme structures, thus refining the identification process.
The TopEC model leverages advanced machine learning techniques, integrating information about distances and angles among atoms to enhance predictive accuracy for enzyme function categorization. Previously, tasks involving enzyme classification often suffered from the pitfalls of fold bias - a scenario wherein the overall shape (fold) interestingly correlates with the enzyme's function. The unique approach taken by TopEC addresses this by using localized three-dimensional descriptors that capture the complex interplay between biochemical characteristics and local shape-dependent features, achieving an impressive F-score of 0.72 in predicting the enzyme commission (EC) numbers.
Traditionally, enzyme classification relied heavily on folding structures that often did not account for subtle variations that significantly affect function. These inconsistencies create considerable obstruction in utilizing databases that contain large volumes of structural data. In response, TopEC utilizes message-passing frameworks to analyze detailed structural elements without introducing fold bias. This allows for robust predictions across a functional space of over 800 EC numbers, encompassing a wide diversity of enzymatic roles.
Understanding enzyme classification and being able to predict it accurately remains critical in various scientific and industrial applications, including drug development and biotechnology. Delving deeper into the mechanisms of enzymatic functions, the TopEC model enables not just more accurate predictions but also opens avenues for discovering new enzymatic activities in proteins previously not classified as enzymes.
Research has shown that while deep learning strategies, particularly graph neural networks, have gained traction in fields like drug discovery and protein interaction prediction, representing protein structures as 3D graphs poses considerable challenges. TopEC rectifies these limitations by creatively using localized 3D descriptors, thus facilitating efficient memory use and improving predictive performance significantly compared to standard GNN methodologies.
What truly distinguishes TopEC from existing models is its capability to operate without prior knowledge of whether a protein functions as an enzyme. This feature amplifies its utility, making it a versatile tool in the enzyme discovery realm, particularly useful when exploring potential functional roles in proteins that might not squarely fall into the traditional classifications. With a biennial increase in genomic data, the ability to seamlessly integrate and identify potential enzymes rapidly distinguishes TopEC.
The research team highlights that through their developed model, understanding the local chemical environment surrounding enzyme reactions allows for a nuanced prediction of enzyme functions. Parameters detailing the local environments feed into the neural network, enhancing its learning process, as TopEC manages to classify enzymes from varying structural backgrounds accurately.
The TopEC framework is not only robust in predictive analytics but also customizable for ongoing or new research endeavors. Published alongside the findings, the modeling framework is hosted on GitHub, encouraging further exploration by the scientific community. Such accessibility ensures that scientists can build upon the modular design of TopEC to develop new function-prediction tools or adapt the architecture for diverse biological applications.
As the scientific community marches forward into the realms of biotechnology, systems biology, and synthetic biology, tools such as TopEC represent a significant advancement in the endeavors toward understanding and engineering enzymes. The importance of these tools lies not just in their immediate applicability but in their potential to pioneer future investigations into the complexities of enzymatic activities and classifications.
This innovation renders a comprehensive look at enzymatic roles, implications for various biochemical applications, and the framework's applicability in future experiments, establishing itself as a pivotal resource for scientists aiming to harness the full potential of enzymes in the wider biological landscape.