The challenge of studying small membrane proteins has always posed obstacles in structural biology, particularly in the context of cryo-electron microscopy (cryo-EM). Recently published research sheds light on innovative strategies to tackle these issues, demonstrating the potential of new biotechnological enhancements that refine our approach to structural analysis.
The study, led by a team of researchers including Ackle, Thavarasah, and Earp, investigates how Legobodies and NabFabs can harmonize with sybodies, a class of small proteins known for their ability to bind to membrane proteins. This innovative study presents that "any sybody can be adapted to the Legobody approach with minimal effort," effectively opening doors for a broader application of cryo-EM to challenging biological targets.
The primary focus of this study is on enhancing cryo-EM resolution when analyzing proteins that are below the 100 kDa threshold, which traditionally face difficulties in achieving clear structural determination due to their low contrast and overall small size. This necessity points to a critical gap in structural biology, where the intricacies of membrane protein dynamics are pivotal to understanding their functionalities in living systems.
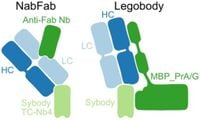
Researchers have long sought methods to engineer small protein domains into loops of the target protein to augment their size. The study highlights the previous use of both single-domain antibodies, or nanobodies, and their engineered derivatives known as sybodies, which can mimic the shape variations found in their natural counterparts. Though nanobodies have shown promise, their moderate size remains a limitation—the continued evolution of tools like Legobodies and NabFabs aims to remedy this shortcoming.
The Legobody approach entails attaching a Fab fragment to the back of a nanobody, connecting it with a protein moiety through a shared helical structure. This modification significantly increases the effective size of the complex, thus enhancing cryo-EM analysis possibilities. Notably, the study emphasizes that "only a subset of sybodies belonging to the loop library can be converted into a format recognized by the NabFab without complementarity-determining region-grafting."
In practical terms, the research unfolds through a series of experiments assessing how various engineered sybodies could be effectively integrated into these new larger assemblies. A new vector called pSBLego was developed, designed to seamlessly incorporate a C-terminal motif that enhances binding efficiency. The results indicate that when expressed from pSBLego, all tested sybodies exhibited binding to Legobody Fab with dissociation constants (KD) measuring between 200 and 350 nM—a favorable result for initiating cryo-EM studies.
Furthermore, the study confirms that by introducing specific amino acid changes, such as the Q108P mutation in loop library sybodies, significant improvements in NabFab binding can be achieved, achieving a KD of approximately 10 nM. In contrast, adjustments made to the concave and convex sybodies yield varying success, indicating a nuanced approach is necessary for modification based on structural characteristics.
This research opens exciting avenues for future studies focused on investigating the structural dimensions of small and complex proteins within membrane environments. The implications extend into drug design and therapeutic development, where understanding the detailed structures of target proteins can lead to the formulation of more effective interventions.
In conclusion, the team’s advancements promise to enhance the scope of cryo-EM applications considerably, particularly concerning small membrane protein studies, while the methodologies set forth encourage widespread adoption in structural biology laboratories globally. As articulated by the researchers, fostering these engineering adaptations not only augments the analysis of membrane proteins but potentially reshapes our perception of their intricate roles and mechanisms in biology, facilitating significant advances in our understanding of cellular functions.