The study reveals temperature-dependent thermodynamic parameters for DNA hybridization, highlighting that assumptions of temperature independence in thermodynamic measures are inaccurate. By employing single-DNA mechanical unzipping experiments combined with calorimetric force spectroscopy, researchers have provided vital insights into how DNA duplex stability varies with temperature.
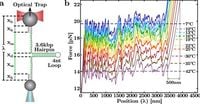
Conducted by a team led by P. Rissone, with contributions from M. Rico-Pasto and Steven B. Smith, the research was published on March 19, 2025. Through novel experimental approaches utilizing a temperature-jump optical trap to unzip a 3,593-base pair DNA hairpin, the researchers explored the energetic landscape of DNA hybridization.
Understanding the thermodynamics of nucleic acid interactions is essential within molecular biology, with implications ranging from PCR amplification methods to gene editing and DNA origami. Historically, many studies assumed that crucial factors such as enthalpy (ΔH) and entropy (ΔS), as well as heat capacity changes (ΔCp), remained constant. However, this study challenges that notion, emphasizing that significant temperature effects can influence DNA stability and hybridization reactions.
The research employed pulling experiments at temperatures ranging from 7 to 42 degrees Celsius, alongside 1M NaCl in a Tris-HCl buffer, observing crucial changes in thermodynamic parameters. Results indicated that the persistence length of DNA increases with temperature, varying from 0.74(7) nm at 280K to 0.88(4) nm at 315K—a roughly 30% increase. This change correlates strongly with the temperature increase, suggesting distinct degrees of freedom within the system during hybridization.
Moreover, the study presented compelling data regarding the inter-phosphate distance of single-stranded DNA (ssDNA), noting a temperature-dependent increase of approximately 5%. During the experiments, the force-distance curves exhibited a pronounced sawtooth pattern, indicating the cooperative unfolding of the DNA segments. This remarkable observation reflects broader patterns in molecular biodegradation and stability that were previously poorly understood.
The authors wrote, "From a microscopic viewpoint, ΔCp relates to the change in the number of degrees of freedom, Δn, in hybridization reactions". This statement underscores the importance of dynamically understanding how molecular interactions evolve with temperature changes. By effectively measuring ΔCp, the researchers reveal that the previously accepted notion of ΔCp = 0 is fundamentally flawed. They found an average heat capacity change of −30(10) cal mol−1 K−1 bp−1, further demonstrating that energetic parameters should indeed be temperature-dependent.
The methodology uniquely combines calorimetric force spectroscopy and machine learning algorithms to estimate thermodynamic parameters with single base pair resolution—a groundbreaking feat. The findings hold particular promise for advancing various biotechnological applications, as greater precision in understanding DNA interactions can dramatically influence gene therapy, design of drugs, and synthetic biology applications.
The researchers noted, "The methodology presented in this work is general and can be straightforwardly extended to other experimental conditions", pointing to future avenues of research that could broaden our understanding of nucleic acids under diverse environmental conditions. The implications of this work extend into realms such as cold denaturation, which raises intriguing considerations about the stability of DNA in extreme environments, possibly informing studies of organisms that thrive in such conditions.
As the research community explores these new parameters, deeper questions will arise regarding the predictive models for DNA behavior at various temperatures. This groundbreaking study is pushing the boundaries of our understanding of nucleic acid thermodynamics, rewriting assumptions that have lingered for too long and paving the way for future insights in DNA interaction research.