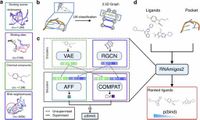
A new deep learning model named RNAmigos2 significantly accelerates virtual screening for RNA-targeted drug discovery, achieving remarkable speed and accuracy compared to traditional methods.
RNA molecules, though smaller in size compared to their protein counterparts, hold immense potential as therapeutic targets. Historically, the focus on proteins for drug development has limited the scope of drug discovery. Recently, a new approach developed by researchers aims to expand this focus by leveraging a deep learning model designed specifically for RNA virtual screening.
RNAmigos2 is a cutting-edge structure-based method that introduces significant efficiencies in identifying RNA-targeting small molecules. By utilizing coarse-grained 3D modeling, synthetic data augmentation, and RNA-specific self-supervision, this method achieves an unprecedented speedup of 10,000 times compared to traditional molecular docking techniques. Importantly, this method ranks active compounds effectively, placing them in the top 2.8% of a structurally diverse test set, indicating its high performance and selective efficacy.
In the quest for effective RNA-targeted therapies, the approval of Risdiplam as the first RNA-targeting drug by the FDA underlines the increasing relevance of RNA in therapeutic development. Researchers emphasize that while RNA molecules, particularly non-coding RNAs, are critical players in numerous biological processes, the drug development landscape has been lacking essential computational tools tailored for RNA.
RNAmigos2 is particularly noteworthy as it not only accelerates the screening process but does so without sacrificing accuracy. The methodologies employed by RNAmigos2 signify a potential turning point in RNA drug discovery. Its development highlights the requirement for a new paradigm that harnesses advanced computational techniques for effective virtual screening in the context of RNA.
By focusing on structure-based approaches, the model capitalizes on the knowledge of RNA 3D structures to search for candidate compounds that could bind effectively. This approach is paramount for discovering new binding modes and mediating high specificity in drug interactions.
The complexities of RNA folding and the unique biophysical phenomena at play present significant challenges for drug development. Consequently, previous computational tools have struggled with the limited availability of RNA structural data in comparison to the wealth of protein data. The RNAmigos2 model uniquely addresses this data shortfall, transforming it into an advantage through the integration of large databases of simulated docking scores and unsupervised training techniques tailored to RNA.
During its evaluation, the RNAmigos2 model was tested against a substantial dataset of 20,000 compounds in a microarray setting. It achieved a mean enrichment factor of 2.93 at a 1% cutoff during screening, reinforcing its viability as a practical tool in drug discovery protocols. Importantly, the model does not merely excel in speed; it efficiently discriminates between active compounds and decoys in a remarkably short period, running its predictions in less than five seconds on a single machine.
Traditional docking methodologies take hours, sometimes reaching up to eight CPU hours per binding site. In stark contrast, RNAmigos2 provides the potential to screen up to 15.4 million compounds within the same time budget dedicated to conventional docking methods. This massive enhancement in efficiency could pave the way for rapid iteration in drug discovery, significantly speeding up the timeline from research to clinical application.
Rising to meet the demands of contemporary pharmaceutical research, RNAmigos2 also demonstrates effectiveness against various binding sites, maintaining performance across diverse RNA structures. Its robustness ensures reliability, even when applied to previously unseen targets and compounds.
The implications of this model extend far beyond basic research. The capacity for rapid and accurate virtual screening enables exploration into uncharted chemical space, opening new avenues for RNA-targeted therapies that could lead to breakthroughs in diseases previously deemed difficult or impossible to treat with traditional protein-targeting approaches.
Looking forward, the development team behind RNAmigos2 plans to refine the model further and is devoted to enhancing its performance through integrating insights from alternative docking technologies. The goal is to create an even more nuanced and effective screening process.
Ultimately, RNAmigos2 represents a substantial leap forward in RNA drug discovery. By making efficient use of computational resources and advancing our understanding of RNA interactions through innovative techniques, this tool could transform how researchers approach RNA-targeting therapeutics.
With a growing commitment to openness, the development team is expected to publicly release the datasets, source code, and model weights, encouraging collaboration within the scientific community. This open-access strategy may catalyze further advancements in the domain and promote a culture of innovation as researchers seek to unlock the potential of RNA-targeted drug development.