Researchers from Zhejiang University have made significant strides in the study of lysine methacrylation, identifying key modification sites on the Cyclophilin A (CypA) protein, which plays a foundational role in various cellular processes. Published on March 17, 2025, their research sheds light on how these modifications influence cellular functions, particularly within redox homeostasis.
Lysine acylation is known as a prevalent form of post-translational modification (PTM), impacting multiple biological mechanisms. While numerous lysine acylation sites have been characterized through heightened analytical methods such as mass spectrometry, researchers previously documented a mere 27 lysine methacrylation (Kmea) sites, largely confined to histone proteins. The current study steps beyond this limitation, establishing the presence of Kmea on the non-histone protein CypA.
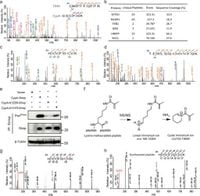
CypA's relevance extends beyond mere enzymatic activity, as it is involved with immunosuppression and has correlations with several human diseases, including cancer. To understand the role of Kmea on this protein, the researchers utilized a groundbreaking genetic code expansion technique to incorporate ε-N-Methacryllysine (MeaK) within target proteins.
Importantly, the modification at site 125 (K125) of CypA was shown to significantly influence cellular redox balance. Specifically, the authors state, "Kmea at CypA site 125 regulates cellular redox homeostasis, and HDAC1 is the regulator of Kmea on CypA." This finding reveals not just the impact of Kmea on protein function, but illuminates the regulatory role played by histone deacetylase 1 (HDAC1) during this process.
Utilizing affinity-purification mass spectrometry (AP-MS), the study identified two lysine residues on CypA (K125 and K131) with Kmea modifications. The researchers confirmed these findings through Western blot analysis, noting substantial variations when these residues were mutated, underscoring the dynamic nature and importance of the Kmea modification.
Along with regulating interactions with oxidoreductase proteins, the researchers also documented the ability of genetically encoded Kmea to undergo methylation, forming ε-N-methyl-ε-N-methacrylation (Kmemea) within living cells. The team’s findings reveal new avenues for inquiry, as they note, "Genetically encoded Kmea can be methylated to form Kmemea in live cells." This points toward novel functional pathways through which CypA might operate, potentially paving the way for future studies on how similar modifications can exert influence in different cellular contexts.
To carry out their experiments, the research team constructed mutants of pyrrolysyl-tRNA synthetase (MbPylRS) to effectively integrate MeaK. Throughout these experiments, they evaluated the efficacy of different constructs, establishing optimal conditions for incorporation, enabling subsequent studies to yield accurate and insightful results.
The results hinted at enhanced interactions, showcasing how 68 proteins uniquely associated with CypA following the incorporation of Kmea, and 10 protein bindings were identified exclusively within CypA K125MeaK experiments. These findings were enlightening not only for molecular biology but for designing targeted therapies which may hinge on the dynamic properties of such modifications.
Finally, these advances open up important dialogues surrounding the dynamic nature of post-translational modifications and their vast roles across cellular landscapes. By demonstrating pathways for studying Kmea beyond histones, the researchers have undoubtedly broadened the horizon for functional discovery within the field. By embedding these molecular insights within the growing body of post-translational modification studies, this research paves the way for future developments and therapeutic targeting of diseases influenced by protein modifications.