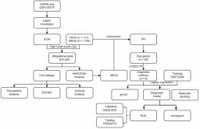
Systemic lupus erythematosus (SLE) is notoriously complex, presenting as one of the most challenging autoimmune diseases due to its heterogeneous manifestations and lack of definitive diagnostic markers. Recent work led by researchers aims to unravel the molecular mechanisms driving this condition, potentially paving the way for improved therapeutic strategies.
A study published on March 17, 2025, utilized microarray datasets from the Gene Expression Omnibus (GEO) database to conduct comprehensive single-cell RNA sequencing (scRNA-seq) analysis. This method led to the identification of 19 distinct cell clusters, from which five major cell types were annotated—CD4 memory T cells, B cells, CD8 effector T cells, dendritic cells, and megakaryocytes.
The researchers focused on mitochondrial-related genes (MRGs) and ferroptosis-related genes (FRGs), scoring the highest expression levels of FRGs in megakaryocytes and MRGs predominantly in B cells. By employing advanced analytical techniques—including pseudotime analysis, cell-cell communication analysis, and Single-Cell Regulatory Network inference and Clustering (SCENIC)—the team delved deep to reveal the heterogeneity present within these cellular populations.
Significantly, the study identified hub genes using high-dimensional weighted correlation network analysis (hdWGNCA) complemented by machine learning algorithms. This approach culminated in the development of a predictive diagnostic model exhibiting high accuracy. The correlation between diagnostic biomarkers and various immune cell types offers new avenues for monitoring and potentially treating SLE.
The analysis also uncovered pivotal interactions within cell populations; for example, enhanced APP_CD74 receptor-ligand interactions were noted between specific megakaryocyte subtypes, particularly PF4 and S100A8. Meanwhile, IGJ and CRIP1 B cells displayed strong MIF_CD74 + CXCR4 interactions. These cellular communications may play significant roles in SLE's immunological dysregulation, providing insights for future therapeutic strategies.
Among the eight diagnostic genes identified were ACTB, ACTG1, CAPZB, FLNA, MYL6, NDUFA1, NDUFB3, and PRDX1. The performance of the diagnostic model was promising, demonstrating area under the curve (AUC) values of 0.919, 0.961, and 0.973 across training, validation, and test cohorts, respectively. This indicates significant potential for these identified markers to guide clinical assessments and management of SLE.
To corroborate these findings, quantitative PCR analyses indicated significant contrasts between SLE patients and control subjects, with PRDX1 expression elevated (p < 0.05) among lupus patients, and lower levels of ACTG1, CAPZB, FLNA, MYL6, and NDUFA1. These markers may prove beneficial for not only diagnosis but for illuminating avenues for new treatment regimens.
The researchers also identified six small molecule drugs capable of interacting with all the diagnostic biomarkers associated with SLE: Acetaminophen, bisphenol A, Doxorubicin, Ivermectin, Tetrachlorodibenzodioxin, and Valproic Acid. Among these, Doxorubicin showed the strongest binding affinity to several proteins integral to the biomarkers, indicating its potential as a therapeutic agent, particularly concerning its impact on mitochondrial function.
Overall, this study provides substantial progress toward elucidation of SLE's underlying mechanisms and highlights the importance of mitochondrial dysfunction and ferroptosis within the pathogenesis of the disease. While promising, these findings call for additional validation through prospective studies encompassing larger patient cohorts to substantiate the clinical applicability of the proposed diagnostic biomarkers and therapeutic strategies.