A large-scale proteomics study reveals causal proteins associated with the risk of developing Inflammatory Bowel Disease, providing new potential therapeutic targets.
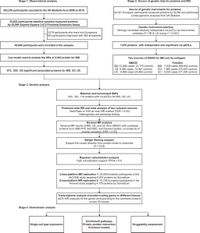
The increasing global prevalence of Inflammatory Bowel Disease (IBD), which includes conditions like Crohn's Disease and Ulcerative Colitis, poses a significant challenge to healthcare systems. A striking analysis from the UK Biobank, incorporating data from over 48,800 IBD-free participants, has uncovered a number of plasma proteins that may play crucial roles in the risk and development of these chronic inflammatory disorders.
IBD affects the quality of life for millions, and early detection, alongside improved treatment methods, remains essential in the management of the disease. Current therapies often yield suboptimal efficacy, leading researchers to explore new avenues in identifying biomarkers that can help forecast disease onset. This innovative study has managed to bridge proteomics and genetics to better understand IBD's underlying mechanisms.
Using data from the UK Biobank Pharma Proteomics Project, the researchers analyzed associations between nearly 3,000 plasma proteins and subsequent IBD development, utilizing advanced statistical methods such as Cox regression analysis. They discovered that 673 proteins were significantly associated with IBD risk, with robust evidence indicating that 18 of these possess causal relationships. Among the highlighted proteins were IL12B and CXCL9, providing insights that not only enhance understanding of IBD's molecular etiology but also spotlight potential therapeutic strategies.
During a median follow-up of 13.6 years, 336 new cases of IBD were documented within the study cohort, primarily consisting of individuals of European ancestry. The findings point towards specific environmental and demographic factors, such as smoking habits and educational attainment, influencing IBD risks, suggesting that these factors must be considered in conjunction with proteomic data when evaluating patient risks.
"By integrating observational and genetic analyses, we identified proteins with significant associations to IBD and its subtypes," stated the authors of the article. Their meticulous methodology not only assessed protein levels at baseline but also examined long-term implications of these proteins on health outcomes, establishing a clearer interpretation of the role of proteins in the disease trajectory.
To fortify their findings, the researchers conducted Mendelian randomization (MR) analyses, a robust genetic technique that minimizes confounding variables. This approach distinguished certain proteins as having a definitive causal impact on IBD risk rather than mere correlation. For example, the presence of IL12B was shown to be significantly linked to increased risk, presenting a new avenue for drug development and intervention.
Furthermore, five proteins, including CD6 and MXRA8, were identified as novel candidates for drug targeting, indicating their potential role in future therapeutic applications. With some proteins already in drug development or being repurposed for related conditions, this research sets the stage for translating scientific discovery into clinical practice.
One of the study's remarkable achievements is its ability to collaborate complex proteomic data with rich genetic information, significantly advancing our comprehension of IBD's complexity. The results reveal promising biomarkers that could be utilized for more precise patient screening and targeted treatment protocols.
"Our findings advance the understanding of IBD’s molecular etiology and highlight potential therapeutic targets," the authors continued, emphasizing the transformative potential of their research in clinical settings. Looking ahead, the identification of these proteins not only offers hope for enhanced patient outcomes but also emphasizes the necessity for continued exploration at the intersection of proteomics and genetic epidemiology.
As the medical community navigates the challenges posed by IBD, the insights from these large-scale studies will undoubtedly aid in the quest for definitive answers and improved therapies. This study demonstrates the power of leveraging vast datasets to extract meaningful insights that can reshape treatment paradigms for chronic diseases.
While the study's strengths lie in its extensive data collection and use of advanced methodologies, the authors also acknowledge certain limitations, including the predominance of European ancestry in the cohort which affects the broader applicability of the findings. Despite this, the groundwork laid by such research signifies a promising step forward in our understanding of IBD and its treatment potential.