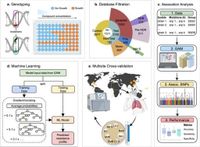
A groundbreaking new method known as the Group Association Model (GAM) is revolutionizing how researchers identify genetic variants linked to drug resistance in microbial infections. Published recently, the study outlines how GAM effectively highlights genes associated with drug resistance, surpassing traditional methods that often produce misleading results.
With the growing challenge of combating microbial drug resistance, particularly in diseases like tuberculosis (TB), this study is timely. Drug-resistant infections, which often require lengthier treatments and lead to increased healthcare costs, represent a significant challenge to public health globally. In fact, an alarming 10.6 million new TB cases were reported in 2021 alone, with over 450,000 of those cases being rifampicin-resistant.
The researchers analyzed a staggering 7,179 Mycobacterium tuberculosis (Mtb) isolates to gauge GAM's performance against the World Health Organization (WHO) mutation catalogue. The results showed that GAM not only provided comparable efficiency in identifying resistance-related genes but did so with significantly fewer false-positive results than the WHO’s traditional methods.
One significant highlight of GAM is its capacity to accurately predict drug resistance in other pathogens too. In a separate analysis involving 3,942 S. aureus isolates, GAM once again proved reliable, showcasing its potential as a versatile tool in microbial genetics.
This study sheds light on the need for innovative methods to tackle the emerging challenge of antimicrobial resistance, emphasizing how current techniques can fall short due to their dependence on catalogued mutations and expert rules. Typical methods, including culture-based techniques and molecular approaches, often take weeks and rely heavily on the experience of researchers to avoid misinterpretations.
The GAM approach identifies genetic variants linked to specific resistance profiles through data clustering of genetically similar isolates, which allows for a more nuanced understanding of the mechanisms behind drug resistance. Moreover, using machine learning (ML) to refine GAM further boosts its predictive accuracy, particularly in datasets containing incomplete information — a common issue in real-world applications.
This innovation is not just applicable to Mycobacterium tuberculosis; the implications for clinical microbiology are profound. As resistance mechanisms evolve, adapting the detection methods to remain ahead becomes crucial. The potential for GAM to seamlessly integrate machine learning techniques represents a formidable advancement in real-time drug susceptibility testing, paving the way for improved treatment strategies for drug-resistant infections.
Overall, the findings from the research underscore a significant leap in our ability to predict drug resistance with higher accuracy, thus promising to transform the management of various infections at a time when the world desperately needs effective methods to combat drug resistance.
As we look to the future, the integration of GAM with machine learning could be a game-changer in the field, enabling healthcare providers to make better-informed decisions in the treatment of drug-resistant infections. Such advancements not only enhance patient outcomes but also curtail the rampant rise of drug-resistant strains on a global scale.
In summary, the development of the Group Association Model could revolutionize the field of drug resistance prediction, providing a clearer, more accurate understanding of the genetic underpinnings driving microbial resistance, ultimately leading to better patient care and management of infectious diseases.