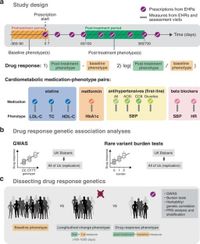
The aim of this study is to shed light on the genetic foundations of individual differences in drug response through the integration of electronic health records and large-scale biobanks, with a particular focus on cardiometabolic medications. By analyzing medication prescription data alongside clinical outcomes, the researchers were able to identify both common and rare genetic variants influencing how effectively patients respond to medications meant to treat conditions like high cholesterol and diabetes.
Genetic variation plays a significant role in the variability seen in responses to medications. Previous research has shown that despite the potential of pharmacogenomics in tailoring drug treatments, there is still a knowledge gap in the field regarding how specific genetic variants affect drug efficacy. The present study sought to fill this gap by examining genetic predictors of drug response via a combination of pharmacogenetic association studies and high-quality clinical data.
Researchers utilized data from the UK Biobank, with over 41,000 participants, along with the All of Us research program, which has approximately 14,000 participants. The study focused on ten significant cardiometabolic drug outcomes, including responses to statins, metformin, and antihypertensive medications. Through employing genome-wide association studies (GWAS) and rare variant burden tests, the researchers unveiled both established and novel correlations linking specific genetic markers to drug efficacy.
The study revealed that the genetic signals associated with drug responses were highly treatment-specific, indicating that genetic factors playing a role in drug metabolism and efficacy vary considerably between patients receiving different therapeutic interventions. Among the significant findings, the analysis confirmed previously known genetic associations with drug responses, such as those involving the APOE gene related to cholesterol metabolism and the SLCO1B1 gene associated with statin response, while also identifying a new association with the GIMAP5 gene linked to HbA1c response to metformin treatment.
Importantly, the results underscore the limitations of conventional methods that often assess drug effects without accounting for genetic variability. For instance, standard polygenic risk scores, which are often used to predict patient responses, explained less than 2% of the variation observed in drug efficacy. These insights suggest a more nuanced understanding of how individual genetic makeup can influence treatment outcomes.
Moreover, this research exemplifies how leveraging electronic health records can enhance pharmacogenomic studies. It emphasizes that employing EHRs in conjunction with biobank data allows researchers to better characterize patient responses in real-world settings, ultimately leading to more personalized medication strategies.
Looking forward, the authors recommend ongoing efforts to combine genetic research with observational data to further elucidate the complexities of drug response. Expanding on this foundation can help realize the promise of personalized medicine, where treatments are tailored based on genetic profiles, enhancing their efficacy while minimizing adverse effects.
In conclusion, this study not only advances our understanding of the genetic underpinnings influencing drug response but also illustrates the potential of integrating data sources to refine pharmacogenomics. By continuing to investigate these genetic associations, researchers may uncover additional pathways for optimizing patient treatment outcomes in cardiometabolic therapeutics.