In a significant advance for antifungal drug development, researchers have uncovered crucial structural differences between the topoisomerase II enzyme of the yeast Saccharomyces cerevisiae and its human counterpart, human topoisomerase IIα (hTopoIIα). This study, published on March 19, 2025, in the journal Scientific Reports, suggests that these variations may pave the way for the creation of selective antifungal agents that could effectively target fungal infections while minimizing impacts on human cells.
Topoisomerases are vital enzymes responsible for managing DNA supercoiling and disentanglement, thus ensuring genomic integrity in all eukaryotic cells. While inhibitors of human topoisomerases have been extensively utilized in cancer chemotherapy, their potential as antifungal agents remains largely unexplored. This study aimed to investigate whether the structural differences between the two types of topoisomerases could allow researchers to develop new drugs specifically targeting the fungal enzyme without affecting human cells.
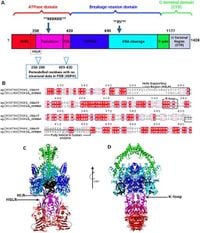
The research team performed a comprehensive sequence analysis of ScTopoII and hTopoIIα, revealing notable differences in the transducer and transducer linker (TDL) domains, as well as in a critical lysine-rich region known as the K-loop. Their findings were bolstered by molecular dynamics simulations, which illustrated these structural differences in action, particularly highlighting unique patterns of hydrophilic and hydrophobic interactions present in ScTopoII that are absent or significantly altered in hTopoIIα.
Phylogenetic analyses further reinforced the significance of these distinctions, demonstrating how the evolutionary paths of fungi and humans have led to divergent structural functions in their topoisomerases. Specifically, the researchers found a 42.9% identity and 63.0% similarity in sequence between the two enzymes, underscoring the potential for selective targeting in drug design.
One of the critical regions of interest in the study was the K-loop in both enzymes. The K-loop of ScTopoII is enriched with lysine residues, which play an essential role in its interactions with DNA during the topoisomerase catalytic cycle. The researchers observed strong hydrogen bond interactions between specific lysine residues and acidic residues nearby, making ScTopoII’s K-loop pivotal for its enzymatic activity. In contrast, hTopoIIα presented very different interactions, influenced predominantly by van der Waals and Coulombic forces.
The study also reported striking differences in the stability of the α-helices and the helix supporting loop regions of the two enzymes. In ScTopoII, the locally structured regions exhibited a more dynamic nature compared to the stable, well-formed helices characteristic of hTopoIIα. This flexibility in ScTopoII might contribute to its unique interaction profile with DNA, which could be exploited to develop targeted inhibitors.
Additionally, researchers used advanced computational tools like AlphaFold to remodel key regions in various organisms’ topoisomerases, scoring their potential for ligand binding. Their investigation identified three promising hotspots for inhibitor development, all situated within or adjacent to the critical K-loop and helix supporting regions in ScTopoII.
Moreover, the study analyzed potential drug binding sites through software assessments and molecular operating environment (MOE) simulations, indicating several unique hotspots in ScTopoII with Drug Scores exceeding 0.6. These discoveries highlight areas where new antifungal compounds could bind selectively, presenting exciting opportunities to treat fungal infections with reduced risk of harm to human cells.
The researchers concluded that their findings not only support the notion that fungal topoisomerases can serve as effective targets for antifungal drug development but also provide a strategic framework for discovering selective inhibitors. They anticipate that further experimental validation of their theoretical findings will lead to the eventual development of new antimicrobial therapies.
This study represents a hopeful stride towards addressing the growing concern of antifungal resistance and the need for novel treatment strategies. By leveraging the inherent differences between human and fungal topoisomerases, scientists are one step closer to developing targeted therapies that could significantly improve patient outcomes while combating resistant fungal strains.