DNA is constantly being damaged by various factors, but it has sophisticated mechanisms to repair itself. A new study reveals significant insights about how single-strand breaks (SSBs) are handled by DNA polymerase beta (Pol β), particularly within the complex structure of chromatin. SSBs occur tens of thousands of times per cell daily and, if left unaddressed, can lead to serious consequences such as mutagenesis, genome instability, and cell death. This study combines biochemical assays and cryogenic electron microscopy (cryo-EM) to illuminate the specific mechanisms by which DNA polymerase beta processes these gaps, contributing to the broader field of DNA repair.
Historically, Pol β has been known as the primary enzyme involved in the repair process of single-strand breaks, especially during base excision repair (BER) and single-strand break repair (SSBR). Until now, the specific way Pol β interacts with nucleosomal DNA to fill 1-nt gaps during these repair processes was poorly understood. The recent findings change this by establishing not just the kinetics of the repair process but also its structural nuances.
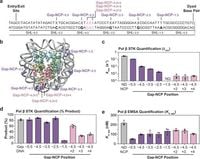
The study began by generating nucleosome core particles containing defined 1-nt gaps. Researchers discovered variations in Pol β's ability to catalyze nucleotide insertion based on the position and orientation of these gaps within the nucleosome, which is the fundamental unit of chromatin structure. Results indicate Pol β utilizes what the authors termed as a 'global DNA sculpting mechanism' to effectively engage with and process these gaps.
Through their research, the team outlined how Pol β binds to the nucleosome, creating substantial structural distortions to access the 1-nt gap. This bending and repositioning of the nucleosomal DNA appears to be dependent on the translational position of the gap—a concept emphasized by the study's kinetics data which showed reduced efficiency for gaps closer to the nucleosome dyad.
Notably, the research also provided cryo-EM structures of Pol β bound to nucleosomal DNA. These images revealed how Pol β interacts with the DNA at the 1-nt gap, identifying the positioning of both the lyase and polymerase domains as it prepares for nucleotide insertion. The findings demonstrated the extensive footprint Pol β has on the nucleosomal DNA, effectively pulling and displacing approximately 35 base pairs of DNA, which is significant for its enzymatic action.
Integration of biochemical data alongside these structural images provides not only clarity on how chromatin influences repair processes but also opens avenues for future research. Understanding how Pol β interacts with chromatin may lead to new insights on genome stability and the mechanisms behind various genetic diseases.
This comprehensive study sheds light on the dynamic and complex nature of DNA repair, showing Pol β's necessity not just as a facilitator of nucleotide insertion but also as a sculptor of the surrounding nucleosomal environment to effectively address single-strand breaks. Given the fundamental role played by Pol β, this study contributes significantly to the field of molecular biology and genetics.