In a groundbreaking study published in March 2025, researchers have benchmarked 13 metagenomic binning tools, uncovering the enhanced efficacy of multi-sample binning strategies in the realm of microbial genomics. This innovative approach has proved pivotal in identifying metagenome-assembled genomes (MAGs) and illuminating critical components within microbial communities, including antibiotic resistance genes and biosynthetic gene clusters.
Microbial ecosystems, vital to Earth's biogeochemical cycles, remain largely unexplored, with a significant part of microbial diversity yet to be characterized. Traditional methods often fall short as many microbes are unculturable. The present study seeks to address this gap by establishing a clear benchmark for evaluating various metagenomic binning tools across differing sequences and approaches.
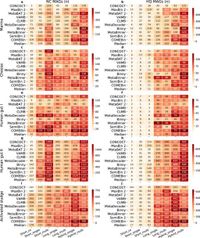
The researchers deployed a comprehensive evaluation of binning tools utilizing short-read, long-read, and hybrid DNA sequencing data. By incorporating diverse methodologies such as co-assembly, single-sample, and multi-sample binning, they aimed to ascertain the most effective approach for recovering high-quality MAGs.
Notably, the study revealed that multi-sample binning outperformed its single-sample counterpart, confirming superior advantages in crucial areas of microbial identification. In benchmark tests, multi-sample binning yielded substantial improvements, recovering 44% more moderate or higher quality (MQ) MAGs from human gut samples, and in marine datasets, it showed a remarkable 100% increase in the recovery of MQ MAGs.
Alongside enhanced recovery rates, multi-sample binning demonstrated its strength in identifying potential antibiotic resistance gene hosts and biosynthetic gene clusters. Specifically, it identified 30%, 22%, and 25% more potential antibiotic resistance gene hosts, alongside 54%, 24%, and 26% more potential biosynthetic gene clusters across short-read, long-read, and hybrid data types, respectively. This finding underscores the technique's potential for advancing microbial risk assessments and drug discovery pursuits.
Veteran researchers H. Han, Z. Wang, and S. Zhu focused not only on identifying which tools delivered optimal performance across various data binning modes but also highlighted which tools ranked best for each combination. For multi-sample approaches, the top performer, COMEBin, demonstrated exceptional results in recovering high-quality MAGs using diverse sequencing methodologies.
Moreover, the ranking assessment showed that while multi-sample binning effectively improved genomic diversity capture, not all binning tools performed equally across all metrics, with some tools being better suited for specific data types. MetaBAT 2, for instance, stood out in terms of runtime efficiency, completing its tasks in just 463 minutes, while still maintaining a lower memory footprint.
While certain top-performing tools excelled in many aspects, it was also noted that the continuous advancement in algorithms presents opportunities for future enhancements in metagenomic binning strategies. With ongoing updates to tools like MAGScoT and DAS Tool, further improvements can be expected on both efficiency and quality of MAGs recovery.
In synopsis, this study presents a crucial step forward in metagenomic research, offering a structured assessment of binning tools that not only aids researchers in selecting optimal methodologies but also pinpoints specific tools capable of facilitating broader microbial diversity discovery. Emphasizing the undeniable need for advancing microbial genomics, the research findings remind us of the significance of enhanced microbial understanding both for addressing current health challenges and for mapping ecological dynamics.
As the field progresses, the call for leveraging multi-sample binning modalities will undoubtedly encourage continuous exploration into previously underrepresented areas of microbial life, thereby augmenting environmental and health-related initiatives worldwide.